Note
Click here to download the full example code
Network Basics
Related MassComposition objects can be managed as a network. In the Process Engineering/Metallurgy disciplines the network will often be called a _flowsheet_.
from copy import deepcopy
from typing import Dict
import pandas as pd
from matplotlib import pyplot as plt
from elphick.mass_composition import MassComposition, Flowsheet
from elphick.mass_composition.mc_node import MCNode
from elphick.mass_composition.datasets.sample_data import sample_data
Create some MassComposition objects
Create an object, and split it to create two more objects.
df_data: pd.DataFrame = sample_data()
obj_mc: MassComposition = MassComposition(df_data, name='Feed')
obj_mc_1, obj_mc_2 = obj_mc.split(0.4, name_1='stream 1', name_2='stream 2')
Placeholder random nodes are created for each MassComposition object. This is done to capture the relationships implicitly defined by any math operations performed on the objects.
for obj in [obj_mc, obj_mc_1, obj_mc_2]:
print(obj.name, obj._nodes)
Feed [324494284532216511557115773226938749275, 126424624233062861302886601543594094071]
stream 1 [126424624233062861302886601543594094071, 132186196820896510251720553826709908991]
stream 2 [126424624233062861302886601543594094071, 220692732893009472278859239807727056057]
Create a Flowsheet object
This requires passing an Iterable of MassComposition objects
fs: Flowsheet = Flowsheet().from_streams([obj_mc, obj_mc_1, obj_mc_2])
Print the node object detail
for node in fs.graph.nodes:
print(fs.graph.nodes[node]['mc'])
{"node_id": "324494284532216511557115773226938749275", "node_name": "Node", "node_subset": "0", "node_type": "SOURCE"}
{"node_id": "126424624233062861302886601543594094071", "node_name": "Node", "node_subset": "0", "node_type": "BALANCE", "balanced": "True"}
{"node_id": "132186196820896510251720553826709908991", "node_name": "Node", "node_subset": "0", "node_type": "SINK"}
{"node_id": "220692732893009472278859239807727056057", "node_name": "Node", "node_subset": "0", "node_type": "SINK"}
Note that the random node placeholder integers have been renumbered for readability.
for obj in [obj_mc, obj_mc_1, obj_mc_2]:
print(obj.name, obj._nodes)
Feed [324494284532216511557115773226938749275, 126424624233062861302886601543594094071]
stream 1 [126424624233062861302886601543594094071, 132186196820896510251720553826709908991]
stream 2 [126424624233062861302886601543594094071, 220692732893009472278859239807727056057]
Print the overall network balanced status
NOTE: presently this only includes node balance status edge balance status will assure the mass-moisture balance is satisfied
print(fs.balanced)
True
Plot the network. Imbalanced Nodes will appear red. Later, Imbalanced Edges will also appear red.
fs.plot()
plt
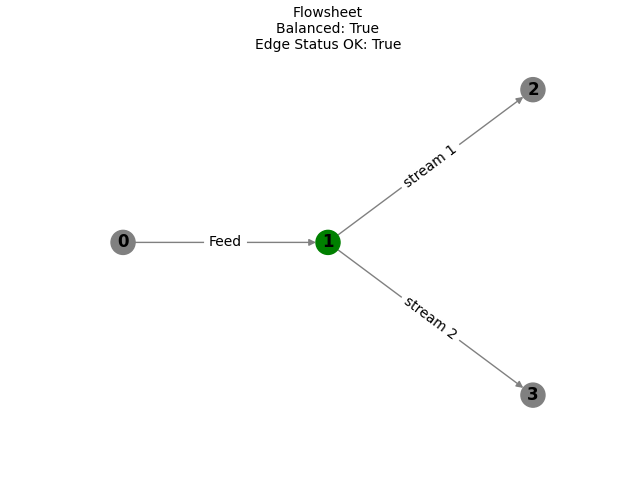
<module 'matplotlib.pyplot' from '/home/runner/work/mass-composition/mass-composition/.venv/lib/python3.10/site-packages/matplotlib/pyplot.py'>
Display the weight averages for all edges (streams) in the network (flowsheet)
df_report: pd.DataFrame = fs.report()
df_report
df_report: pd.DataFrame = fs.report(apply_formats=True)
df_report
Plot the interactive network using plotly
fig = fs.plot_network()
fig
Plot the Sankey
fig = fs.plot_sankey()
fig
Demonstrate the table-plot
fig = fs.table_plot(plot_type='sankey', table_pos='top', table_area=0.3)
fig
fig = fs.table_plot(plot_type='network', table_pos='bottom', table_area=0.3)
fig
Expand the Network with Math Operators
obj_mc_3, obj_mc_4 = obj_mc_2.split(0.8, name_1='stream 3', name_2='stream 4')
obj_mc_5 = obj_mc_1.add(obj_mc_3, name='stream 5')
fs2: Flowsheet = Flowsheet().from_streams([obj_mc, obj_mc_1, obj_mc_2, obj_mc_3, obj_mc_4, obj_mc_5])
fig = fs2.table_plot(plot_type='sankey', table_pos='left')
fig
Setting Node names
nodes_before: Dict[int, MCNode] = fs.nodes_to_dict()
print({n: o.node_name for n, o in nodes_before.items()})
{0: 'Node', 1: 'Node', 2: 'Node', 3: 'Node'}
fs.set_node_names(node_names={0: 'node_0', 1: 'node_1', 2: 'node_2', 3: 'node_3'})
nodes_after: Dict[int, MCNode] = fs.nodes_to_dict()
print({n: o.node_name for n, o in nodes_after.items()})
{0: 'node_0', 1: 'node_1', 2: 'node_2', 3: 'node_3'}
Setting Stream data
First we show how to easily access the stream data as a dictionary
stream_data: Dict[str, MassComposition] = fs.streams_to_dict()
print(stream_data.keys())
dict_keys(['Feed', 'stream 1', 'stream 2'])
We will replace stream 2 with the same data as stream 1.
new_stream: MassComposition = deepcopy(fs.get_edge_by_name('stream 1'))
# we need to rename to avoid a creating a duplicate stream name
new_stream.name = 'stream 1 copy'
fs.set_stream_data({'stream 2': new_stream})
print(fs.streams_to_dict().keys())
dict_keys(['Feed', 'stream 1', 'stream 1 copy'])
Of course the network is now unbalanced as highlighted in the Sankey
fig = fs.table_plot()
fig
Methods to modify relationships
Sometimes the network that is automatically created may not be what you are after - for example flow may be in the wrong direction. We’ll learn how to modify an existing network, by picking up the network above.
Let’s break the links for the _stream 1_.
fs.reset_stream_nodes(stream="stream 1")
fig = fs.table_plot()
fig
We’ll now break all remaining connections (we could have done this from the start).
fs.reset_stream_nodes()
fig = fs.table_plot()
fig
Now we’ll create some linkages - of course they will be completely rubbish and not balance.
fs.set_stream_parent(stream="stream 1", parent="Feed")
fs.set_stream_child(stream="stream 1", child="stream 1 copy")
fig = fs.table_plot()
fig
Perhaps less useful, but possible, we can build relationships by setting nodes directly.
fs.reset_stream_nodes()
fs.set_stream_nodes(stream="stream 1", nodes=(1, 2))
fs.set_stream_nodes(stream="stream 1 copy", nodes=(2, 3))
fig = fs.table_plot()
fig
Total running time of the script: ( 0 minutes 2.194 seconds)