Note
Go to the end to download the full example code.
Load Block Model
Demonstrates loading a block model in parquet format into pyvista.
import logging
from pathlib import Path
import numpy as np
import pandas as pd
import pyvista as pv
from elphick.geomet import Sample
from elphick.geomet.block_model import BlockModel
logging.basicConfig(level=logging.DEBUG)
Load
block_model_filepath: Path = Path("block_model_copper.parquet")
# Load the parquet file into a DataFrame
df = pd.read_parquet(block_model_filepath)
print(df.shape)
df.head()
(1689600, 1)
Create a BlockModel
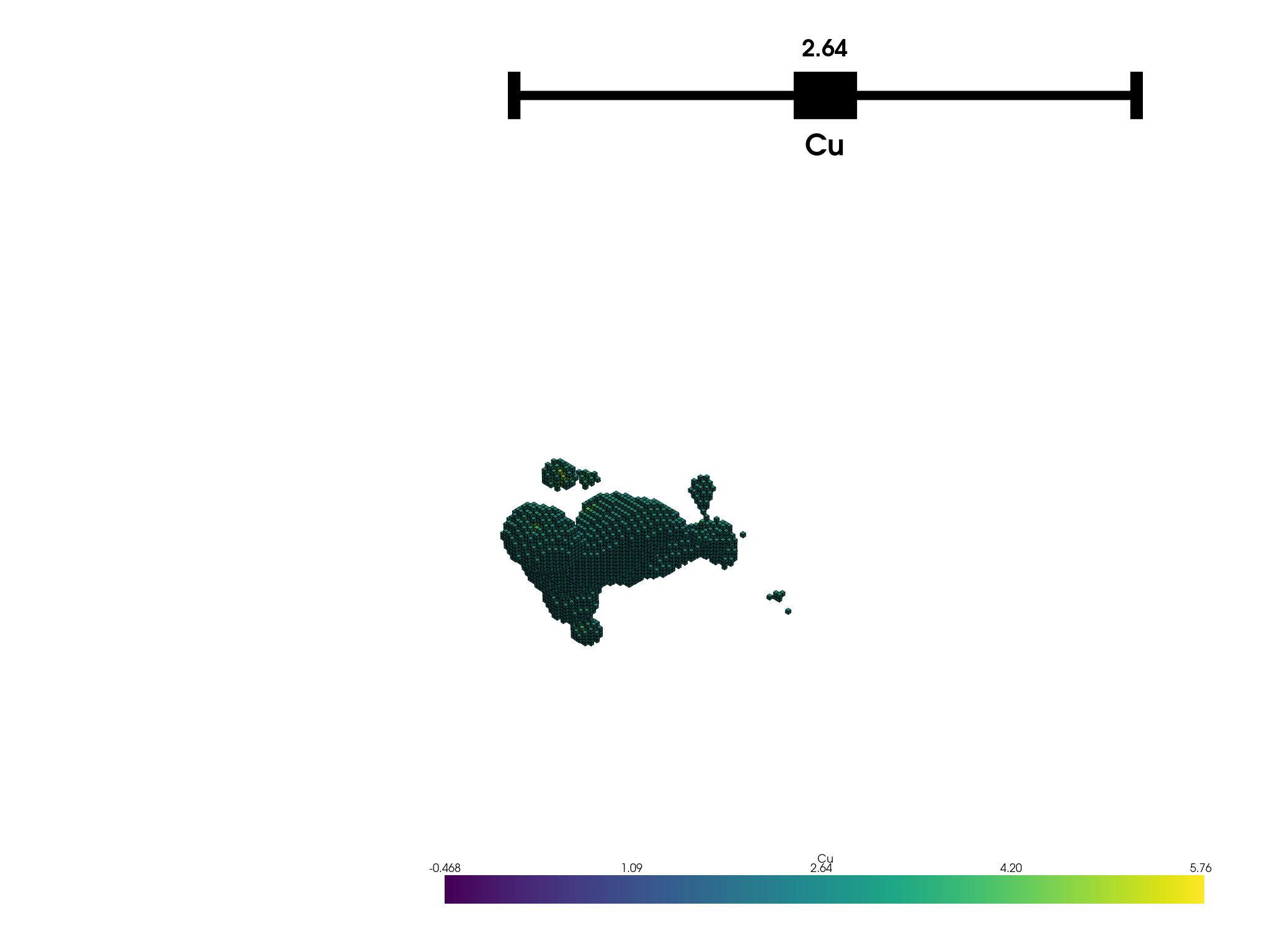
The BlockModel class is a subclass of MassComposition and inherits all its attributes and methods. The block model plotted below is regular, that is, it has a record for every block in the model. Blocks are the same size and adjacent to each other. The block model is created from a DataFrame that has columns for the x, y, z coordinates and the copper percentage.
We need to assign a dry mass (DMT) to the block model to conform to the underlying MassComposition class.
bm: BlockModel = BlockModel(data=df.rename(columns={'CU_pct': 'Cu'}).assign(**{'DMT': 2000}),
name='block_model', moisture_in_scope=False)
bm._mass_data.head()
print(bm.is_irregular)
print(bm.common_block_size())
False
(np.float64(10.0), np.float64(10.0), np.float64(10.0))
bm.data.head()
Plot the block model
bm.plot('Cu').show(auto_close=False)
Filter the data
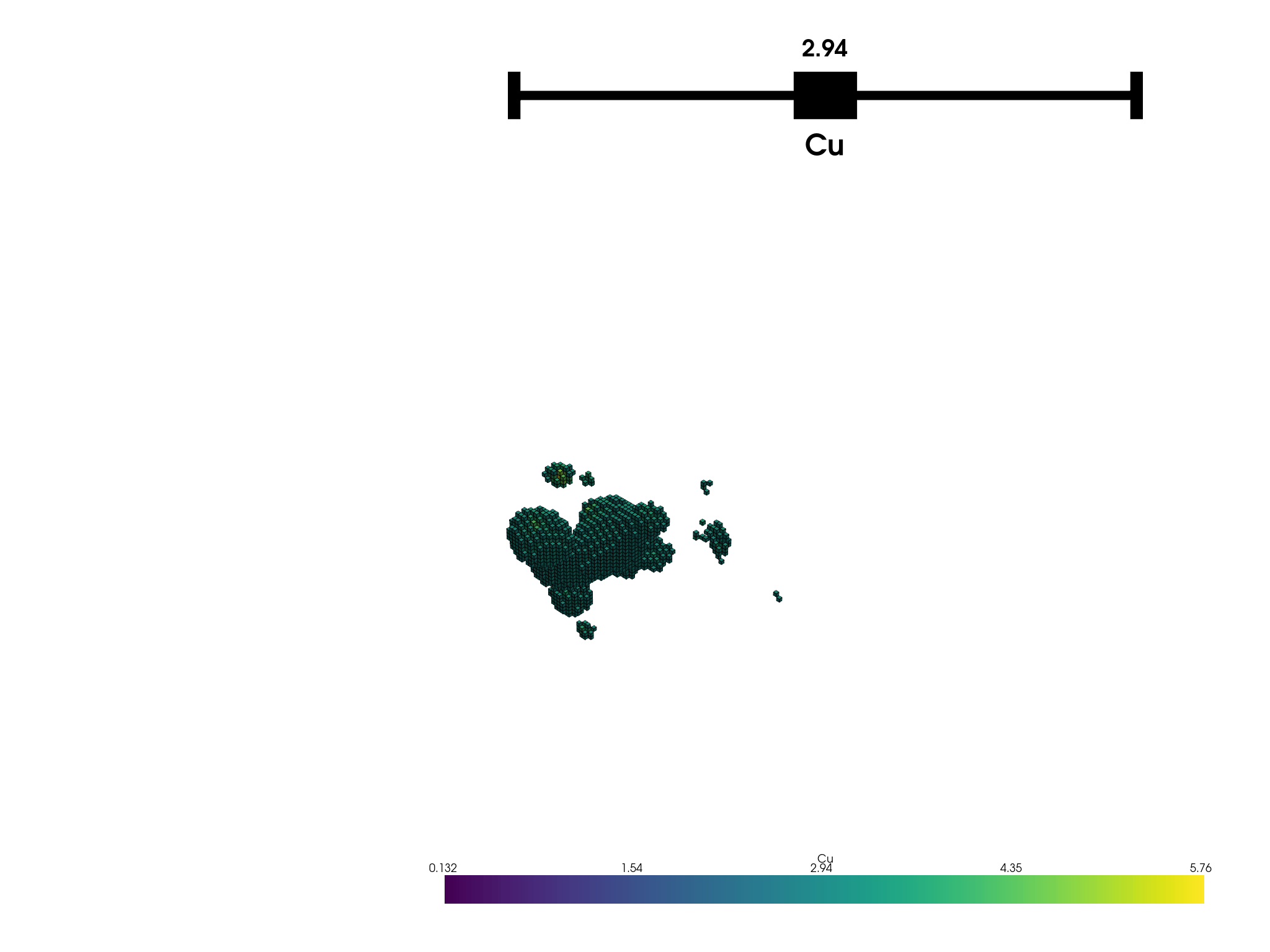
When a dataframe that represents a regular block model (a record for every block) is filtered, the resulting block model cannot be regular anymore. This is because the filtering operation may remove blocks that are adjacent to each other, resulting in a block model that is irregular. This example demonstrates this behavior. The plot below is generated from a filtered block model that was originally regular.
df_filtered = df.query('CU_pct > 0.132').copy()
bm2: BlockModel = BlockModel(data=df_filtered.rename(columns={'CU_pct': 'Cu'}).assign(**{'DMT': 2000}),
name='block_model', moisture_in_scope=False)
bm2._mass_data.shape
(766492, 2)
bm2.plot('Cu').show(auto_close=False)
Total running time of the script: (0 minutes 2.887 seconds)